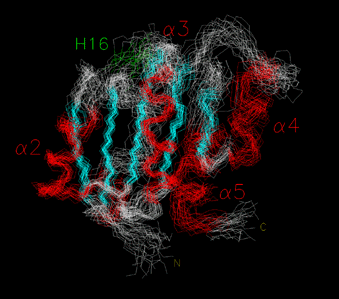
NMR STRUCTURE OF THE B12-BINDING (MUTS) DOMAIN OF GLUTAMATE MUTASE
A number of B12-enzymes are now known that contain a conserved B12-binding domain homologous to MutS. One sequence motif, Asp-x-His-x-x-Gly, stands out as being invariant in all these enzymes. The crystal structures of two of these enzymes, methylmalonyl-CoA mutase and the MeCbl-binding domain of methionine synthase have been solved. They show that the MutS-like domain binds the lower face of the corrin ring and the nucleotide "tail" of the coenzyme and that the conserved histidine residue coordinates to the central cobalt atom.
In collaboration with Prof. Bernhard Krautler's group in Innsbruck, we have used NMR to determine the solution structure of MutS in the absence of coenzyme. The overall structure is very similar to that of the B12-binding domains of methylmalonyl-CoA mutase and methionine synthase. However, the loop containing the Asp-x-His-x-x-Gly motif and the helix following it, that is also predicted to make contacts with coenzyme, are in an extended, mobile conformation. This suggests that the protein and coenzyme fold around each other upon binding, and provides the first clues as to assembly of the protein:coenzyme complex. This work was recently published in Structure.