Research
The Zhang lab is most interested in the relative roles of chance and necessity in evolution. There are two major research areas.
I. Yeast as an experimental system for studying evolution.
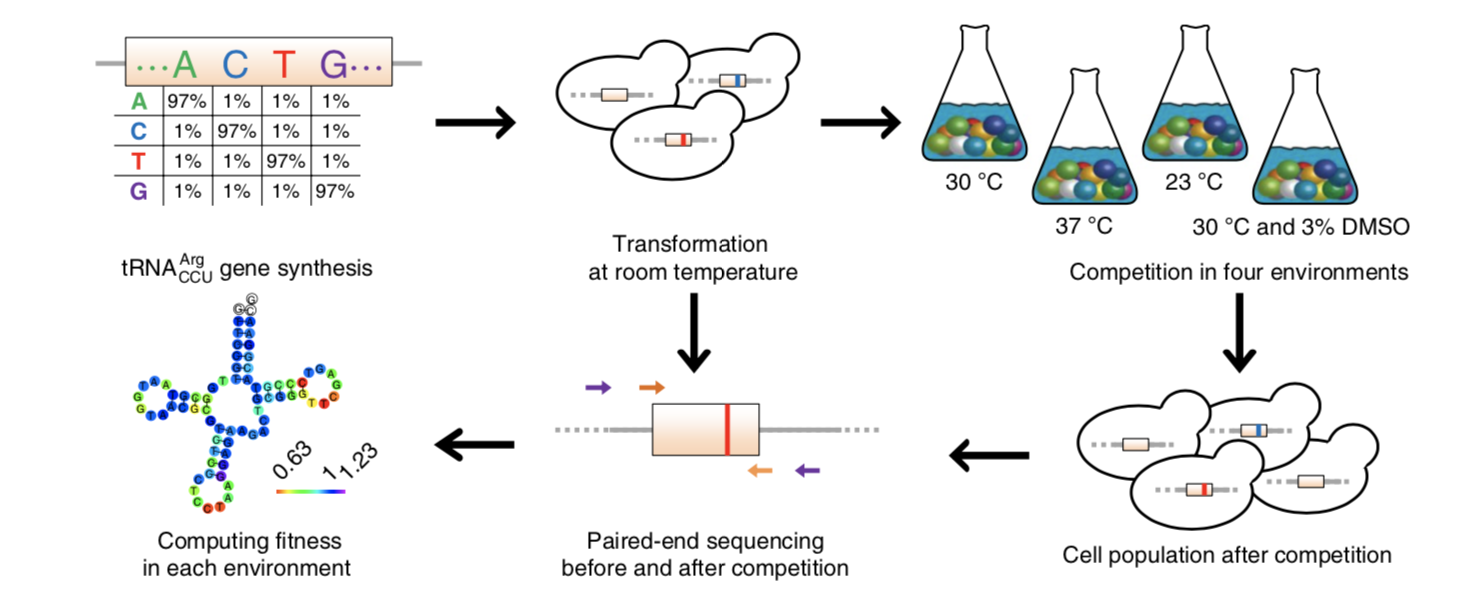
We are using the budding yeast Saccharomyces cerevisiae and its relatives as model organisms to understand a variety of evolutionary processes, including questions about (i) the rate and molecular spectrum of mutation in different environments, (ii) fitness effects of synonymous, nonsynonymous, and regulatory mutations, (iii) epistasis and fitness landscapes, (iv) impacts of pleiotropy on molecular evolution, (v) position effects on gene expression level and expression noise, and (vi) evolutionary origin and entrenchment of new genetic systems. We use a variety of experimental approaches, including, for example, experimental evolution, mutation accumulation, genome/transcriptome sequencing, single-cell sequencing, and genome editing.

II. Computational evolutionary genomics.
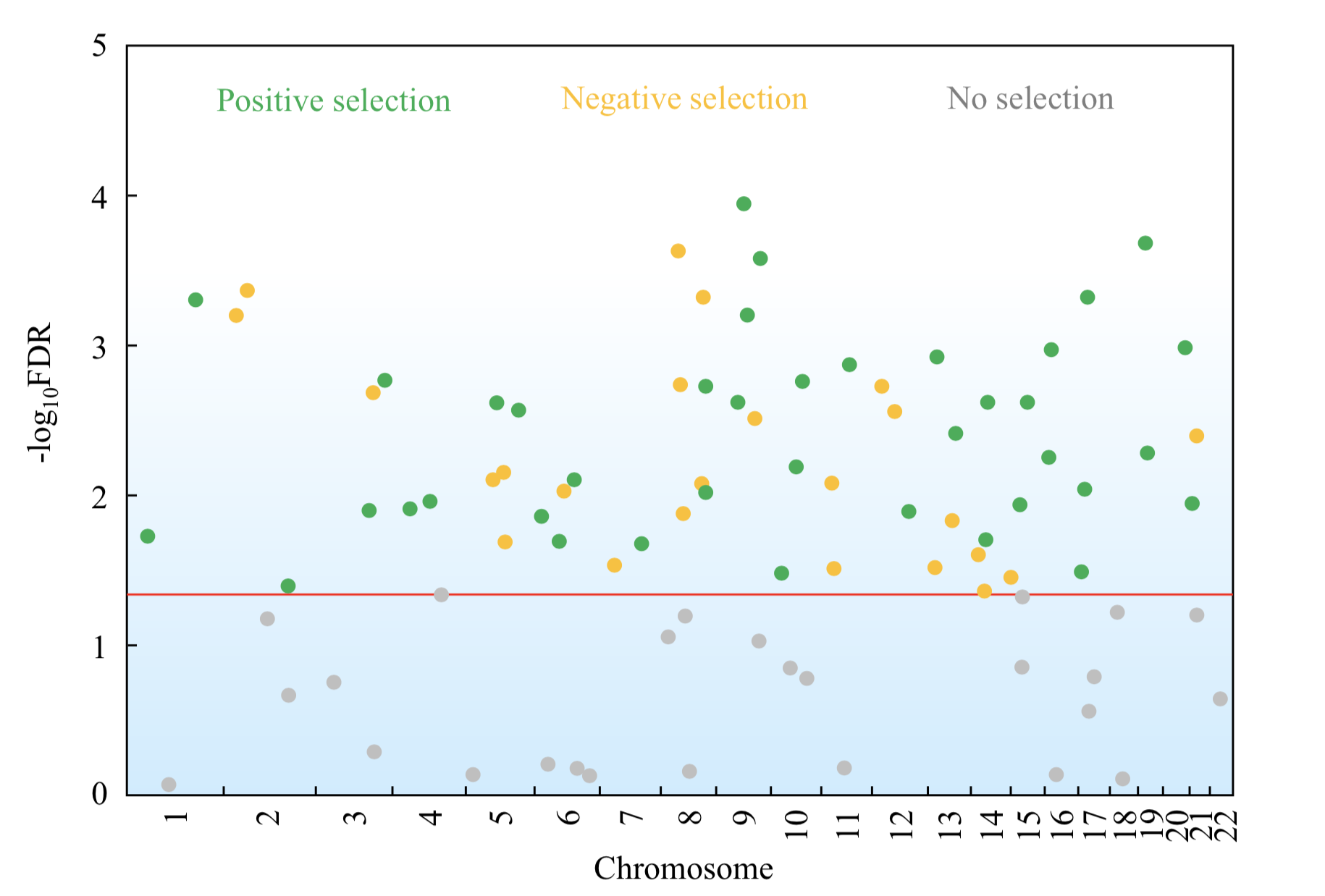
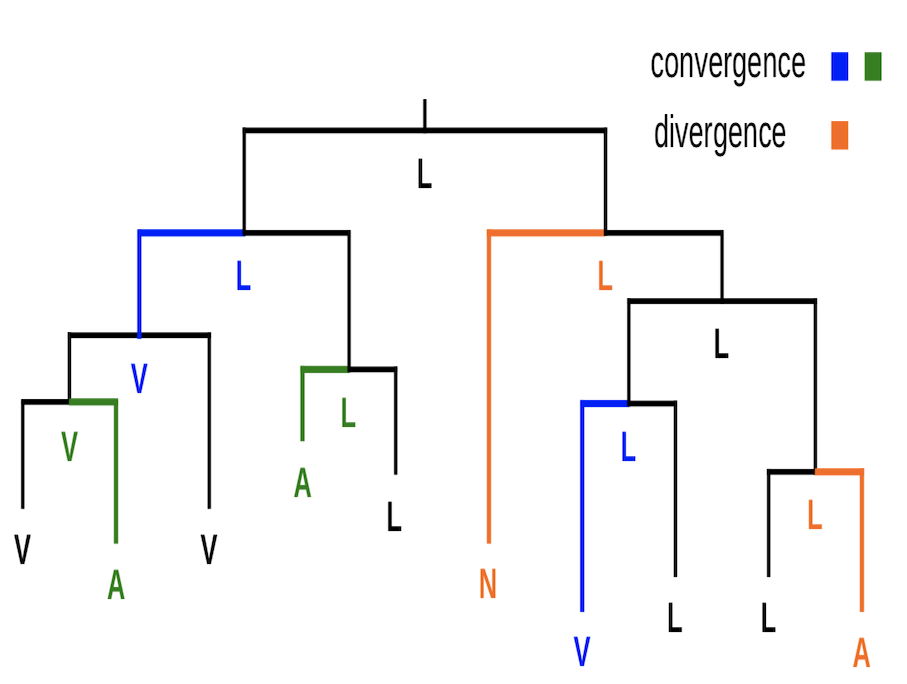
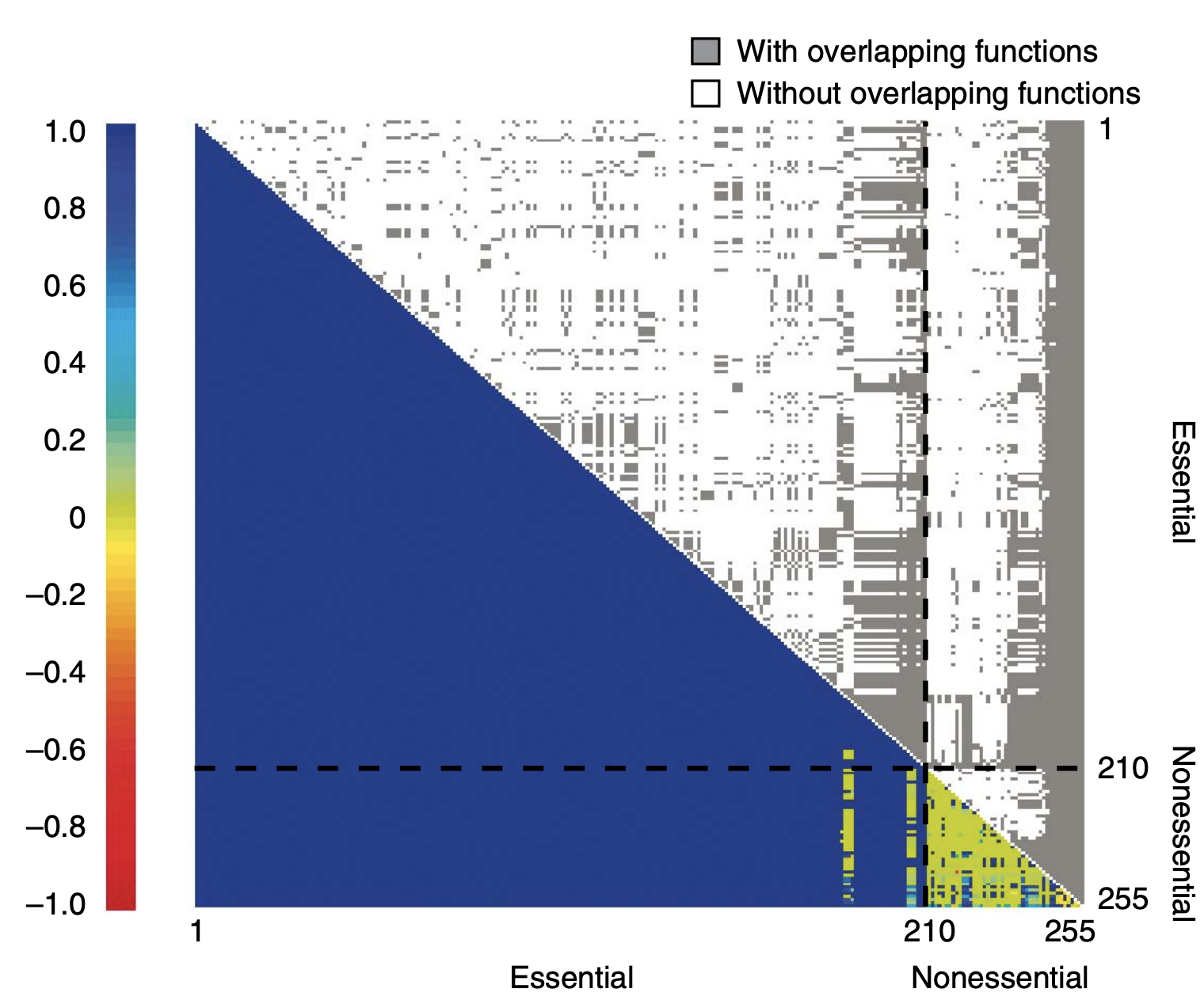
Using evolutionary, genomic, and/or systemic approaches, we analyze publicly available data to characterize and understand pleiotropy, epistasis, fitness landscapes, gene-environment interaction, phenotypic plasticity, adaptation, gene expression level and noise, posttranscriptional modification, translational regulation, origin of new genes, mechanisms of DNA and protein sequence evolution, methods for detecting molecular adaptation, molecular phylogenetic methods, evolutionary medicine, and other important subjects in genetics and evolution. We are not limited in the study organism in this line of research, but most often analyze functional genomic data from yeasts and mammals. Projects may also involve modeling and simulation.
Students and postdoctoral fellows are strongly encouraged to develop their own experimental or computational projects in the general area of evolutionary genetics/genomics.