How do we Sequence DNA?
DNA Sequencing is at the center of the Human Genome Project, which promises to revolutionize
the Biomedical Sciences and the treatment of human diseases. This page is designed to help you
understand how DNA is sequenced.
If you are looking for information on our DNA sequencing service facility, our home page is here:
The University of Michigan DNA Sequencing Core
|
First you need to know a few key terms:
As you go through the subsequent discussion, you may need to jump back here to
refresh your memory on various definitions.
- DNA
- We assume you've read through the
description of DNA structure,
an earlier link in this thread ... right? You hopefully also read the link that describes
DNA Denaturation, Annealing and Replication,
since the following page builds on those basics.
- Plasmid
- A 'plasmid' is a small, circular piece of DNA that is often found in bacteria.
This innocuous molecule might help the bacteria survive in the presence of
an antibiotic, for example, due to the genes it carries. To scientists, however,
plasmids are important because (i) we can isolate them in large quantities,
(ii) we can cut and splice them, adding whatever DNA we choose, (iii) we can
put them back into bacteria, where they'll replicate along with the bacteria's
own DNA, and (iv) we can isolate them again - getting billions of copies of
whatever DNA we inserted into the plasmid! Plasmid are limited to sizes of
2.5-20 kilobases (kb), in general.
- BAC
- The term 'BAC" is an acronym for 'Bacterial Artificial Chromosome', and in principle,
it is used like a plasmid. We construct BACs that carry DNA from humans or mice or
wherever, and we insert the BAC into a host bacterium. As with the plasmid, when we
grow that bacterium, we replicate the BAC as well. Huge pieces of DNA can be easily
replicated using BACs - usually on the order of 100-400 kilobases (kb). Using BACs,
scientists have cloned (replicated) major chunks of human DNA. This, as you will see
later, is critical to the Human Genome Project.
- Vector
- The 'vector' is generally the basic type of DNA molecule used to replicate your DNA,
like a plasmid or a BAC.
- Insert
- The 'insert' is a piece of DNA we've purposely put into another (a 'vector') so that
we can replicate it. Usually the 'insert' is the interesting part, consequently. In
the case of the Human Genome Project or other sequencing projects, the insert is
the part we want to sequence - the part we don't know. Usually we know the complete
DNA sequence of the vector.
- Shotgun Sequencing
- Shotgun sequencing is a method for determining the sequence fo a very large piece of
DNA. The basic DNA sequencing reaction can only get the sequence of a few hundred
nucleotides. For larger ones (like BAC DNA), we usually fragment the DNA and insert
the resultant pieces into a convenient vector (a plasmid, usually) to replicate them.
After we sequence the fragments, we try to deduce from them the sequence of the
original BAC DNA.
- For more definitions ...
- See our Molecular Biology Glossary.
Now for the details on how DNA Sequencing works:
|
|
|
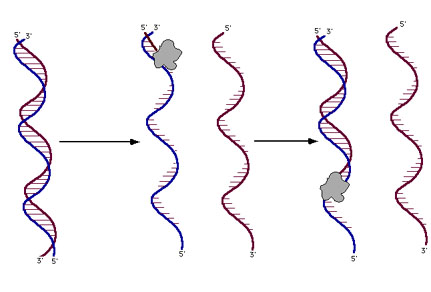
DNA sequencing reactions are just like the PCR reactions for replicating DNA
(refer to the previous page DNA Denaturation, Annealing and Replication).
The reaction mix includes the template DNA, free nucleotides,
an enzyme (usually a variant of Taq polymerase) and a 'primer' - a small piece
of single-stranded DNA about 20-30 nt long that can hybridize to one strand
of the template DNA.
The reaction is initiated by heating until the two strands of DNA separate, then
the primer sticks to its intended location and DNA polymerase starts elongating
the primer. If allowed to go to completion, a new strand of DNA would be the
result. If we start with a billion identical pieces of template DNA, we'll get
a billion new copies of one of its strands.
|
|
|
|
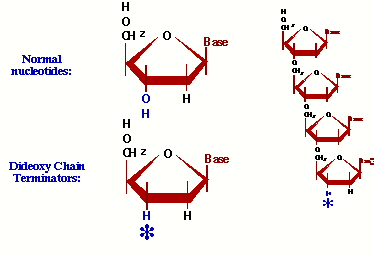
Dideoxynucleotides: We run the reactions, however, in the presence of a dideoxyribonucleotide. This
is just like regular DNA, except it has no 3' hydroxyl group - once it's added
to the end of a DNA strand, there's no way to continue elongating it.
Now the key to this is that MOST of the nucleotides are regular ones, and just a fraction
of them are dideoxy nucleotides....
|
|

|
|
Replicating a DNA strand in the presence of dideoxy-T
MOST of the time when a 'T' is required to make the new strand, the enzyme will
get a good one and there's no problem. MOST of the time after adding a T, the
enzyme will go ahead and add more nucleotides. However, 5% of the time, the enzyme will
get a dideoxy-T, and that strand can never again be elongated. It eventually breaks away
from the enzyme, a dead end product.
Sooner or later ALL of the copies will get terminated by a T, but each time the
enzyme makes a new strand, the place it gets stopped will be random. In millions
of starts, there will be strands stopping at every possible T along the way.
ALL of the strands we make started at one exact position. ALL of them end with
a T. There are billions of them ... many millions at each possible T position.
To find out where all the T's are in our newly synthesized strand, all we have
to do is find out the sizes of all the terminated products!
|
|
|
|
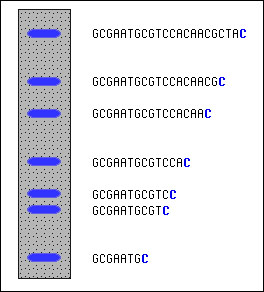
Here's how we find out those fragment sizes.
Gel electrophoresis can be used to separate the fragments by size and measure
them. In the cartoon at left, we depict the results of a sequencing reaction
run in the presence of dideoxy-Cytidine (ddC).
First, let's add one fact: the dideoxy nucleotides in my lab have been chemically
modified to fluoresce under UV light. The dideoxy-C, for example, glows blue. Now
put the reaction products onto an 'electrophoresis gel' (you may need to refer to 'Gel Electrophoresis' in
the Molecular Biology Glossary), and you'll see something like depicted
at left. Smallest fragments are at the bottom, largest at the top. The positions
and spacing shows the relative sizes. At the bottom is the smallest fragment that's
been terminated by ddC; that's probably the C closest to the end of the primer (which
is omitted from the sequence shown). Simply by scanning up the gel, we can see that
we skip two, and then there's two more C's in a row. Skip another, and there's
yet another C. And so on, all the way up. We can see where all the C's are.
|
|

|
Putting all four deoxynucleotides into the picture:
Well, OK, it's not so easy reading just C's, as you perhaps saw in the last figure.
The spacing between the bands isn't all that easy to figure out. Imagine,
though, that we ran the reaction with *all four* of the dideoxy nucleotides
(A, G, C and T) present, and with *different* fluorescent colors on each. NOW
look at the gel we'd get (at left). The sequence of the DNA is rather obvious
if you know the color codes ... just read the colors from bottom to top: TGCGTCCA-(etc).
(Forgive me for using black - it shows up better than yellow).
|
|
|
|
An Automated sequencing gel:
That's exactly what we do to sequence DNA, then - we run DNA replication reactions
in a test tube, but in the presence of trace amounts of all four of the
dideoxy terminator nucleotides. Electrophoresis is used to separate the resulting
fragments by size and we can 'read' the sequence from it, as the colors march past
in order.
In a large-scale sequencing lab, we use a machine to run the electrophoresis step
and to monitor the different colors as they come out. Since about 2001, these
machines - not surprisingly called automated DNA sequencers - have used 'capillary
electrophoresis', where the fragments are piped through a tiny glass-fiber capillary
during the electrophoresis step, and they come out the far end in size-order.
There's an ultraviolet laser built into the machine that shoots through the liquid
emerging from the end of the capillaries, checking for pulses of fluorescent
colors to emerge. There might be as many as 96 samples moving through as many
capillaries ('lanes') in the most common type of sequencer.
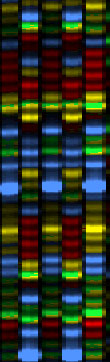
At left is a screen shot of a real fragment of sequencing gel (this one from an
older model of sequencer, but the concepts are identical). The four colors red,
green, blue and yellow each represent one of the four nucleotides.
The actual gel image, if you could get a monitor large enough to see it all at
this magnification, would be perhaps 3 or 4 meters long and 30 or 40 cm wide.
|
|
|
A 'Scan' of one gel lane:
We don't even have to 'read' the sequence from the gel - the computer does that for us!
Below is an example of what the sequencer's computer shows us for one sample. This is a plot of
the colors detected in one 'lane' of a gel (one sample), scanned from smallest fragments
to largest. The computer even interprets the colors by printing the nucleotide
sequence across the top of the plot. This is just a fragment of the entire file,
which would span around 900 or so nucleotides of accurate sequence.
The sequencer also gives the operator a text file containing just the nucleotide sequence,
without the color traces.
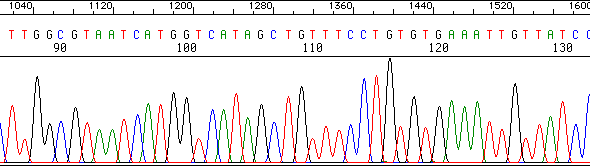
|
As you have seen, we can get the sequence of a fragment of DNA as long as
900 or so nucleotides. Great! But what about longer pieces? The human genome is
3 *billion* bases long, arranged on 23 pairs of chromosomes. Our sequencing
machine reads just a drop in the bucket compared to what we really need!
To do it, we break the entire genome up into manageable
pieces and sequence them. There are two approaches currently in use:
- The Publically-funded Human Genome Project:
The National Institutes of Health and the National Science Foundation
have funded the creation of 'libraries' of BAC clones. Each BAC carries
a large piece of human genomic DNA on the order of 100-300 kb. All of
these BACs overlap randomly, so that any one gene is probably on several
different overlapping BACs. We can replicate those BACs as many times as necessary, so
there's a virtually endless supply of the large human DNA fragment.
In the Publically-funded project, the BACs are subjected to shotgun sequencing
(see below) to figure out their sequence. By sequencing all the BAC's, we
know enough of the sequence in overlapping segments to reconstruct how the original
chromosome sequence looks.
- A Privately-Funded Sequencing Project: Celera Genomics
An innovative approach to sequencing the human genome has been pioneered by
Celera Genomics. The founders of this company realized that it might be
possible to skip the entire step of making libraries of BAC clones. Instead,
they blast apart the entire human genome into fragments of 2-10 kb and
sequence those. Now the challenge is to assemble those fragments of sequence
into the whole genome sequence.
Imagine, for example that you have hundreds of 500-piece puzzles, each being
assembled by a team of puzzle experts using puzzle-solving computers. Those
puzzles are like BACs - smaller puzzles that make a big genome manageable.
Now imagine that Celera throws all those puzzles together into one room and
scrambles the pieces. They, however, have scanners that scan all the puzzle
pieces and huge computers that figure out where they all go.
It is controversial still as to whether the Celera approach will succeed on
a puzzle as large as the human genome. Whether it does or not, they have
certainly stirred up the intellectual pot a bit.
|
Shotgun sequencing: assembly of random sequence fragments
To sequence a BAC, we take millions of copies of it and chop them all up randomly.
We then insert those into plasmids and for each one we get, we grow lots
of it in bacteria and sequence the insert. If we do this to enough fragments,
eventually we'll be able to reconstruct the sequence of the original BAC
based on the overlapping fragments we've sequenced!
|
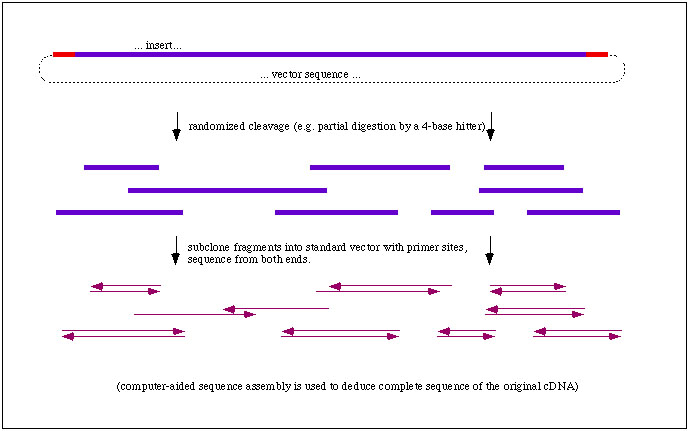
|
|
|