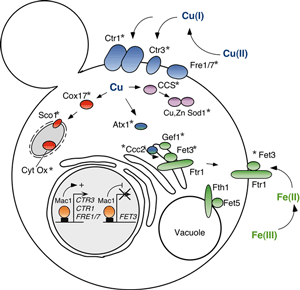
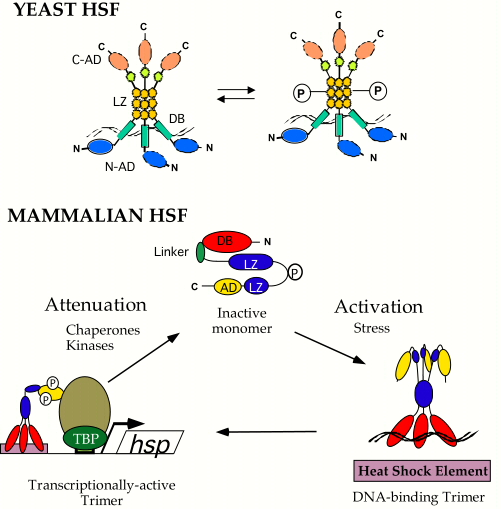
| My colleagues and I are interested in how organisms sense and respond to specific physiological and environmental changes. One area focuses on the mechanisms by which heat shock protein (Hsp) expression is induced in response to stress, pathophysiological conditions, pharmacological agents and normal cell growth and differentiation. Hsps are a class of highly conserved proteins that function in protein folding, assembly, targeting, processing and degradation and, as such, are often elevated in their levels in response to the stresses associated with growth and disease. Heat Shock Factors (HSFs) are a family of highly conserved transcription factors that respond to stressful conditions to activate the expression of Hsp genes. As shown in the figure to the right, the baker’s yeast HSF exists in cells largely as a DNA bound homotrimer which, in response to stress, is hyperphosphorylated and transcriptionally active. Although yeast HSF is activated by multiple, distinct stresses, our results suggest that different signal transduction pathways and protein kinases communicate with HSF in response to disctinct stresses. We are interested in delineating these pathways to understand how cells use a single transcription factor to respond to multiple stresses. Humans and mice have multiple distinct HSF isoforms that appear to respond to unique signals and activate the expression of distinct target genes. As shown in the figure to the right, under non-stressful conditions human HSF1 (hHSF1) exists largely as a monomer unable to bind DNA with high affinity, which is thought to be sequestered through intramolecular interactions across coiled-coil domains, through its linker domain, and through inter-molecular interactions with some of the Hsps themselves. In response to stessful conditions, hHSF1 is thought to form homotrimers which bind to Hsp gene promoters, is hyperphosphorylated and transcriptionally competent. We have developed a powerful yeast-based genetic system to dissect out many of the important details for hHSF1 regulation, to identify novel proteins that regulate hHSF1 and to understand how distinct mammalian HSF isoforms activate different target genes. To learn more about our studies on Heat Shock Factors, please see some of our recent publications. |
 |